
Dr. Richard C. Willson
Huffington-Woestemeyer ProfessorProfessor of Biochemical & Biophysical Sciences
Our laboratory works on applications of biomolecular recognition to bioseparations, molecular diagnostics and process analytical technology. We collaborate productively with groups in a broad range of disciplines. Over time, our group members have had expertise in chemical, biochemical and biomedical engineering, biochemistry, chemistry, chemical biology, computer science, materials science, app development, molecular genetics, microbiology, and nanofabrication.
Research Faculty:
- Katerina Kourentzi, Research Professor
- Binh V. Vu, Research Associate Professor
2025 Fellow, Royal Society of Chemistry
2021 ACS Alan Michaels Award in Bioseparations
2020 Esther Farfel Award (highest faculty award of the University of Houston; for research, service and teaching)
2018-present Co-PI, FDA Western National Pediatric Device Consortium
2017-present Diagnostics co-lead, CDC Western Gulf Center for Vector-Borne Diseases
2015 Pierce Award in Affinity Technology of the International Society for Molecular Recognition (first US awardee in over 20 years)
2015 Fellow, National Academy of Inventors
2014 Fellow, American Chemical Society
2014 Elected to Phi Kappa Phi
2013-present Huffington-Woestemeyer Professor
2012 Co-organizer, AACC Oak Ridge Conference on Emerging Diagnostics, San Jose, CA
2011-present Fellow, American Association for the Advancement of Science
2010-2013 John and Rebecca Moores Professor (campus-wide competitive)
2009 Fluor-Daniel Award (highest faculty award of the UH College of Engineering)
2008-2015 Theme Leader, Diagnostics, NIH Western Regional Center of Excellence
2005 UH Cullen College of Engineering Senior Faculty Research Award
2003-present President, past President, Council, International Society for Molecular Recognition
2001 van Lanen Award, Biochemical Technology Division, American Chemical Society
1999-present Fellow, American Institute of Medical and Biological Engineering
1999 Chair, ACS Division of Biochemical Technology
1990-1995 NSF Presidential Young Investigator
2008-present: Editorial Board, PLOS ONE
2008-present: Affiliate, Quantitative & Computational Biosciences Program, Baylor College of Medicine
2007-present: Senior Affiliate Member, The Methodist Hospital Research Institute
2006-present: Member, Faculty of 1000
2004-2007: President, International Society for Molecular Recognition (Program of 2005 and 2007 meetings)
2003-present: Editorial board, Biotechnology Progress
2001-present: Editorial board, Journal of Molecular Recognition
1999: Chair, American Chemical Society; Division of Biochemical Technology
1999: Member/Past Chair, UH Intellectual Property Committee
1998-2002: Council on Chemical Research (CCR) Vision 2020 Roadmap committee on Bioprocessing
Selected Publications
- Brosamer K, Kourentzi K, Willson RC, Vu BV, Glowstick-inspired smartphone-readable reporters for sensitive, multiplexed lateral flow immunoassays. Communications Engineering 2, 31-36, 2023
- Danthanarayana AN, Nandy S, Kourentzi K, Vu B, Shelite TR, Travi BL, Brgoch J, Willson RC. , Smartphone-readable RPA-LFA for the high-sensitivity detection of Leishmania kDNA using nanophosphor reporters. PLoS Negl Trop Dis.17(7):e0011436. doi: 10.1371/journal.pntd.0011436, 2023
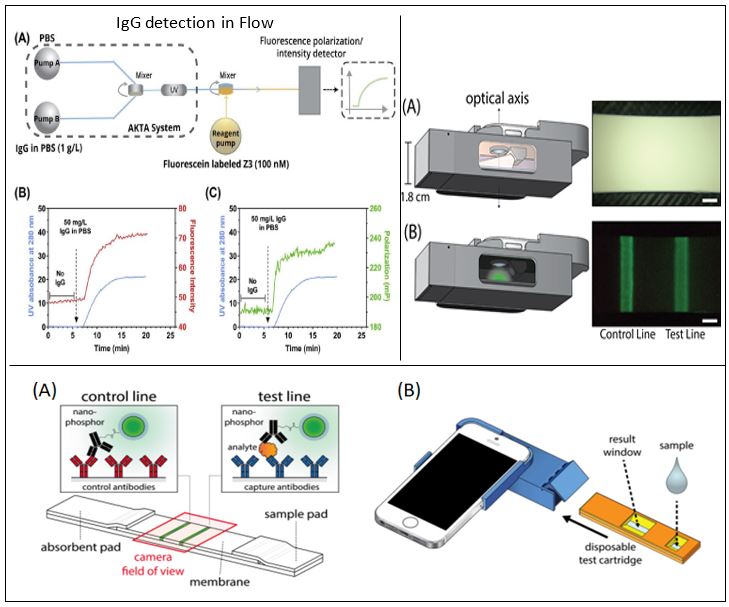
- Goyal A, Vu B, Maranholkar V, Patil U, Kourentzi K, Willson RC, Continuous monitoring of IgG using immobilized fluorescent reporters. Biotechnol Bioeng.120(2):482-490. doi: 10.1002/bit.28254, 2023
- Nandy S, Maranholkar VM, Crum M, Wasden K, Patil U, Goyal A, Vu B, Kourentzi K, Mo W, Henrickson A, Demeler B, Sen M, Willson RC, Expression and Characterization of Intein-Cyclized Trimer of Staphylococcus aureus Protein A Domain Z. Int J Mol Sci. 24(2):1281. doi: 10.3390/ijms24021281, 2023, 2023
- Vu BV, Brosamer K, McDaniel N, Kourentzi, K, Willson RC, Fernando H, “Glow ELISA”: sensitive immunoassay with minimal equipment and stable reagents. Analyst, Advance Article, doi: 10.1039/d3an01623d, 2023
- Lei R, Vu B, Kourentzi K, Soomro S, Danthanarayana AN, Brgoch J, Nadimpalli S, Petri M, Mohan C, Willson RC, A novel technology for home monitoring of lupus nephritis that tracks the pathogenic urine biomarker ALCAM. Front Immunol. 13:1044743. doi: 10.3389/fimmu.2022.1044743, 2022
- Goux HJ, Raja B, Kourentzi K, Trabuco JRC, Vu BV, Paterson AS, Kirkpatrick A, Townsend B, Lee M, Truong VTT, Pedroza C, Willson RC., "Evaluation of a nanophosphor lateral-flow immunochromatographic assay for self-testing for herpes simplex virus type 2 seropositivity." PLoS One. 14(12):e0225365. doi: 10.1371/journal.pone.0225365, 2019
- Paterson AS, Raja B, Mandadi V, Townsend B, Lee M, Vu B, Brgoch J, Willson RC, A low-cost smartphone-based platform for highly sensitive point-of-care testing with persistent luminescent phosphors. Lab on a Chip. 17:1051-1059, 2017
- Kisley L, Chen J, Mansur AP, Shuang B, Kourentzi K, Poongavanam MV, Chen WH, Dhamane S, Willson RC, Landes CF,, Unified superresolution experiments and stochastic theory provide mechanistic insight into protein ion-exchange adsorptive separations. Proc Natl Acad Sci 111: p. 2075-2080., 2014
Research partially sponsored by:


- Añez-Lingerfelt M, Fox GE, Willson RC., “Reduction of DNA contamination in RNA samples for reverse transcription-polymerase chain reaction using selective precipitation by compaction agents,” Anal Biochem., 384(1):79-85, 2009
- Mohan S, Kourentzi K, Schick KA, Uehara C, Lipschultz CA, Acchione M, Desantis ME, Smith-Gill SJ, Willson RC., “Association Energetics of Cross-Reactive and Specific Antibodies,” Biochemistry, 2009
- Potty AS, Kourentzi K, Fang H, Jackson GW, Zhang X, Legge GB, Willson RC., “Biophysical characterization of DNA aptamer interactions with vascular endothelial growth factor,”Biopolymers., 91(2):145-56, 2009
- Zhang X, Potty AS, Jackson GW, Stepanov V, Tang A, Liu Y, Kourentzi K, Strych U, Fox GE, Willson RC., “Engineered 5S ribosomal RNAs displaying aptamers recognizing vascular endothelial growth factor and malachite green,” J Mol Recognit., 22(2):154-61, 2009
- Zuo G,Roberts DJ,Lehman SG,Jackson GW,Fox GE, Willson RC., “Molecular assessment of salt-tolerant, perchlorate- and nitrate-reducing microbial cultures,” Water Sci Technol.,60(7):1745-1756, 2009
- Añez M, Putonti C, Fox GE, Fofanov Y, Willson RC., “Exhaustive computational identification of pathogen sequences far-distant from background genomes: Identification and experimental verification of human-blind dengue PCR primers,” J Biotechnol.,133(3):267-76, 2008
- Basile LJ, Willson RC, Sewell BT, Benedik MJ., “Genome mining of cyanide-degrading nitrilases from filamentous fungi,” Appl Microbiol Biotechnol., 80(3):427-35, 2008
- Jackson GW, McNichols RJ, Fox GE, Willson RC., “Toward Universal Flavivirus Identification by Mass Cataloging,” J Mol Diagn., 10(2):135-141, 2008
- Kourentzi K, Srinivasan M, Smith-Gill SJ, Willson RC., “Conformational flexibility and kinetic complexity in antibody-antigen interactions,” J Mol Recognit., 21(2):114-21, 2008
- Nick Taylor J, Darugar Q, Kourentzi K, Willson RC, Landes CF., “Dynamics of an anti-VEGF DNA aptamer: a single-molecule study,” Biochem Biophys Res Commun., 373(2):213-8, 2008
- Peterson KJ, Sadowsky JD, Scheef EA, Pal S, Kourentzi KD, Willson RC, Bresnick EH, Sheibani N, Gellman SH., “A fluorescence polarization assay for identifying ligands that bind to vascular endothelial growth factor,” Anal Biochem., 378(1):8-14, 2008
- Vu BV, Litvinov D, Willson RC., “Gold nanoparticle effects in polymerase chain reaction: favoring of smaller products by polymerase adsorption,” Anal Chem., 80(14):5462-7, 2008
- Cano T, Offringa N, Willson RC., “The effectiveness of three multi-component binding models in describing the binary competitive equilibrium adsorption of two cytochrome b(5) mutants,” J Chromatogr A., 1144(2):197-202, 2007
- Fu JY, Balan S, Potty A, Nguyen V, Willson RC., “Enhanced protein affinity and selectivity of clustered-charge anion-exchange adsorbents,” Anal Chem., 79(23):9060-5, 2007
- Jackson GW, McNichols RJ, Fox GE, and Willson RC., “Universal bacterial identification by mass spectrometry of 16S ribosomal RNA cleavage products,” International Journal of Mass Spectrometry, 261(2-3):218-226, 2007
- Larios-Sanz M, Kourentzi KD, Warmflash D, Jones J, Pierson DL, Willson RC, Fox GE., “16S rRNA beacons for bacterial monitoring during human space missions,” Aviat Space Environ Med., 78(4 Suppl):A43-7, 2007
- Ortego BC, Whittenton JJ, Li H, Tu SC, Willson RC., “In vivo translational inaccuracy in Escherichia coli: missense reporting using extremely low activity mutants of Vibrio harveyi luciferase,” Biochemistry, 46(48):13864-73, 2007
- Svitel J, Boukari H, Van Ryk D, Willson RC, Schuck P., “Probing the functional heterogeneity of surface binding sites by analysis of experimental binding traces and the effect of mass transport limitation,” Biophys J., 92(5):1742-58, 2007
- Tucker DL, Ott CM, Huff S, Fofanov Y, Pierson DL, Willson RC, Fox GE., “Characterization of Escherichia coli MG1655 grown in a low-shear modeled microgravity environment,” BMC Microbiol., 7(1):15, 2007
- Fu JY, Potty AS, Fox GE, Willson RC., “Water-elutability of nucleic acids from metal-chelate affinity adsorbents: enhancement by control of surface charge density,” J Mol Recognit.,19(4):348-53, 2006
- Jackson GW, McNichols RJ, Fox GE, Willson RC., “Bacterial genotyping by 16S rRNA mass cataloging,” BMC Bioinformatics, 7:321, 2006
- Murphy JC, Cano T, Fox GE, Willson RC., “Compaction agent protection of nucleic acids during mechanical lysis,” Biotechnol Prog., 22(2):519-22, 2006
- Potty AS, Fu JY, Balan S, Haymore BL, Hill DJ, Fox GE, Willson RC., “Neutral additives enhance the metal-chelate affinity adsorption of nucleic acids: role of water activity,” J Chromatogr A., 1115(1-2):88-92, 2006
- Putonti C, Chumakov S, Mitra R, Fox GE, Willson RC, and Fofanov Y., “Human-blind probes and primers for dengue virus identification,” FEBS J., 273(2):398-408, 2006
- Zhang Z, Jackson GW, Fox GE, Willson RC., “Microbial identification by mass cataloging,”BMC Bioinformatics, 7:117, 2006
- Cano T, Murphy JC, Fox GE, & Willson R.C., “Separation of Genomic DNA from Plasmid DNA by selective renaturation with IMAC capture,” Biotechnology Progress, 21: 1472-1477, 2005
- Cano T, Offringa ND, and Willson RC, “Competitive ion-exchange adsorption of proteins: isotherms with controlled competitor concentration,” J Chromatogr A., 24;1079(1-2):116-26, 2005
- Chumakov, S., Belapurkar, C., Putonti, C., Li, T.-B., Pettitt, B.M., Fox, G.E., Willson, R.C. and Fofanov, Y., “Theoretical Basis for Universal Identification Systems for Bacteria and Viruses,” Journal of Biological Physics and Chemistry, 5(4); 121-128, 2005
- Cohen, G.H., Silverton, E.W., Padlan, E.A., Dyda, F., Wibbenmeyer, J.A., Willson, R.C., and Davies, D.R., “Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 Å resolution: comparison with results from isothermal titration calorimetry,” Acta Crystallographica Section D: Biological Crystallography, D61, 628-633, 2005
- Jackson GW, and Willson R, “Preparative electrophoresis with on-column optical fiber monitoring and direct elution into a minimized volume,” Biotechnol Lett., 27(22):1739-43, 2005
- Jandhyala DM, Willson RC, Sewell BT, and Benedik MJ, “Comparison of cyanide-degrading nitrilases,” Appl Microbiol Biotechnol., 68(3):327-35, 2005
- Mulder BA, Anaya S, Yu P, Lee KW, Nguyen A, Murphy J, Willson R, Briggs JM, Gao X, and Hardin SH, “Nucleotide modification at the gamma-phosphate leads to the improved fidelity of HIV-1 reverse transcriptase,” Nucleic Acids Res., 33(15):4865-73, 2005
- Tucker DL, Karouia F, Wang J, Luo Y, Li TB, Willson RC, Fofanov Y, and Fox GE, “Effect of an artificial RNA marker on gene expression in Escherichia coli,” Appl Environ Microbiol.,71(7):4156-9, 2005
- Forsman, Z.H., Lednicky, J.A., Fox, G.E., Willson, R.C., White, Z.S., Halvorson, S.J., Wong, C., Lewis, A.M. Jr, and Butel, J.S., “Phylogenetic analysis of polyomavirus simian virus 40 from monkeys and humans reveals genetic variation,” Journal of Virology, 78(17):9306-16, 2004
- Balan, S. E., Murphy, J. C., Galaev, I., Kumar, A., Fox, G. E., Mattiasson, B., and Willson, R. C., “Metal Chelate Affinity Precipitation of Nucleic Acids,” Biotechnology Letters, 25:1111-1116, 2003
- DeWalt, B., Murphy, J. C., Fox, G. E., and Willson, R. C., “Compaction Agent Clarification of Microbial Lysates,” Protein Expression and Purification, 28(2) 220 – 223, 2003
- D’Souza, L.M., Larios-Sanz, M., Setterquist, R. A., Willson, R. C., and Fox, G. E., “Small RNA Sequences are readily stabilized by inclusion in a carrier rRNA,” Biotechnology Progress,19: 734-738, 2003
- Gibbs, P.R., Riddle, R.R., Marchal, M., Benedik, M.J. and Willson, R.C., “Purification and characterization of 2'aminobiphenyl-2,3-diol 1,2-dioxygenase from Pseudomonas sp. LD2,” Protein Expression and Purification, 32:35-43, 2003
- Gibbs, P.R., Uehara, C.S., Nguyen, P.T., and Willson, R.C., “Imaging Polarimetry for High Throughput Chiral Screening,” Biotechnology Progress, 19(4); 1329-1334, 2003
- Jandhyala, D., Berman, M., Meyers, P.R., Sewell, B.T., Willson, R.C., and Benedik, M.J., “CynD, the Cyanide Dihydratase from Bacillus pumilus: Gene Cloning and Structural Studies,” Applied and Environmental Microbiology, 69:4794-4805, 2003
- Kourentzi, K.D., George E. Fox, G.E. and Willson. R.C., “Hybridization-responsive fluorescent DNA probes containing the adenine analog 2-aminopurine,” Analytical Biochemistry, 322(1), 124-126, 2003
- Murphy J.C., Fox G.E., and Willson, R.C., “Enhancement of anion-exchange chromatography of DNA using compaction agents,” J. Chromatogr. A., 984: 215-21, 2003
- Murphy, J. C., Jewell, D. L., White, K. I., Fox, G. E., and Willson, R. C., “Nucleic Acid Separations Utilizing Immobilized Metal Affinity Chromatography,” Biotechnology Progress, 19: 982-986, 2003
- Riddle, R. R., Gibbs, P. R., Willson, R.C., and Benedik, M. J., “Recombinant carbazole-degrading strains for enhanced petroleum processing,” J. Ind. Microbiol. Biotechnol., 30: 6-12, 2003
- Riddle, R., P.R. Gibbs, R.C. Willson and M.J. Benedik, “Purification and Properties of 2-hydroxy-6-oxo-6-(2'-aminophenyl)hexa-2,4-dienoic acid Hydrolase Involved with Microbial Degradation of Carbazole,” Protein Expression and Purification, 28:182-189, 2003
- Kourentzi, K.D., G.E. Fox and R.C. Willson, “Microbial Identification by Immunohybridization Assay of Artificial RNA Labels,” J. Microbiol. Methods, 49(3), 301-306, 2002
- Murphy, J.C., G.E. Fox and R.C. Willson, “Compaction Agents Enhance Anion-Exchange Adsorption of Nucleic Acids,” Journal of Chromatography, 2002
- Shick, K.A., S. Mohan, S.J. Smith-Gill and R.C. Willson, “Calorimetric Study of Monoclonal Antibody HyHEL-10 Association with Hen and Japanese Quail-Egg Lysozymes,” Biochimica et Biophysica Acta, 2002
- Zhang, Z., R.C. Willson and G.E. Fox, “Identification of Characteristic Oligonucleotides in the 16S Ribosomal RNA Sequence Dataset,” Bioinformatics, 18, 244-250, 2002
- Kourentzi, K.D., G.E. Fox and R.C. Willson, “Microbial Detection with Low-Molecular-Weight RNA,” Current Microbiology, 43(6), 444-447, 2001
- Kourentzi, K.D., G.E. Fox and R.C. Willson, “Rapid Identification of Microorganisms using 5S rRNA Specific Molecular Beacons,” Current Microbiology, 43, 444, 2001
- Murphy, J.C., G.E. Fox and R.C. Willson, “RNA Isolation and Fractionation using Compaction Agents,” Analytical Biochemistry, 295, 143, 2001
- D’Souza, L.M., R.C. Willson and G.E. Fox, “Expression of Marker RNAs in Pseudomonas Putida,” Current Microbiology, 40, 91-95, 2000
- K.A. Xavier, S.M. McDonald, J.A. McCammon, and R.C. Willson, “Association and dissociation kinetics of Bobwhite quail lysozyme with monoclonal antibody HyHEL-5,” Protein Engineering, 12(1), 79-83, 1999
- Li, H., Ortego, B.C., Maillard, K.I., Willson, R.C., and Tu, S.-C., “Effects of mutations of the alpha His-45 residue of Vibrio harveyi luciferase on the yield and reactivity of flavin peroxide intermediate,” Biochemistry, 38(14), 4409-4415, 1999
- Murphy, J.C., Wibbenmeyer, J.A., Fox, G.E. and Willson, R.C., “Purification of plasmid DNA using selective precipitation by compaction agents,” Nature Biotechnology, 17, 822, 1999
- Riddle, R.R., Willson, R.C., and Benedik, M.J., “Temperature- and solvent-tolerant mutants of filamentous bacteriophage helper M13 KO7,” Biotechnology Letters, 21(1):87-90, 1999
- Singh, N. and Willson, R.C., “Boronate affinity adsorption of RNA: possible role of conformational changes,” Journal of Chromatography, 840, 205-213, 1999
- Wibbenmeyer, J.A., P. Schuck, S.J. Smith-Gill, and R.C. Willson, “Salt Links Dominate Affinity of Antibody HyHEL-5 for Lysozyme Through Enthalpic Contributions,” Journal of Biological Chemistry, 274(38), 26838-42, 1999
- Wibbenmeyer, J.A., Xavier, K.A., Smith-Gill, S.J. and Willson, R.C., “Cloning, Expression, and Characterization of the Fab Fragment of the Anti-Lysozyme Antibody HyHEL-5,” BBA, 1430, 191-202, 1999
- Benedik, M.J., Gibbs, P.R., Riddle, R.R., and Willson, R.C., “Microbial denitrogenation of fossil fuels,” Trends Biotechnol., 16, 390-395, 1998
- Xavier, K.A., Shick, K.A., and Willson, R.C., “Association and Dissociation kinetics of anti-hen egg lysozyme monoclonal antibodies HyHEL-5 and HyHEL-10,” Biophys. J., 74, 2036-2045, 1998
- Shick, K.A., Xavier, K.A., Rajpal, A., Smith-Gill, S.J. and Willson, R.C., “Association of the anti-hen egg lysozyme monoclonal antibody HyHEL-5 with avian species variant and mutant lysozymes,” Biochim. Biophys. Acta, 1340, 205-214, 1997
- Xavier, K.A., Shick, K.A., Smith-Gill, S.J. and Willson, R.C., “Involvement of water molecules in the association of monoclonal antibody HyHEL-5 with Bobwhite quail lysozyme,” Biophys. J., 73, 2116-2125, 1997
- Chacko, S., Silverton, E.W., Smith-Gill, S.J., Davies, D.R., Shick, K.A., Xavier, K.A., Willson, R.C., Jeffrey, P.D., Chang, C.Y.Y., Sieker, L.C., and Sheriff, S., “Refined structures of Bobwhite quail lysozyme uncomplexed and complexed with the HyHEL-5 Fab fragment,” Proteins: Structure, Function, and Genetics, 26, 55-65, 1996
- Maillard, K.I., Benedik, M.J. and Willson, R.C., “Rapid detection of mutagens by induction of luciferase-bearing prophages in E. coli,” Environmental Science and Technology, 30(8), 2478-2483, 1996
- Moates, F.C., Somani, M., Annamalai, J., Richardson, J.T, Luss, D. and Willson, R.C., “Infrared Thermographic Screening of Combinatorial Libraries of Heterogeneous Catalysts,” Industrial and Engineering Chemistry Research, 35(12), 4801-4803, 1996