Faculty

Undergraduate Program Director
Joint Appointments in Biomedical Engineering and Biology & Biochemistry
Research in the Cirino lab generally aims to improve “biocatalysis” (broadly, enzyme- and/or microbial-based conversion of chemical feedstocks into higher value chemical products). This includes advancing production of renewable fuels and chemicals, and repurposing or otherwise harnessing often complex natural chemical transformations orchestrated by one or more enzymes. This is accomplished through protein and metabolic engineering, and synthetic biology applications. Most notably, we have pioneered the customization and use of bacterial transcription factors as genetically-encoded biosensors/molecular reporters for high-throughput biocatalyst screening. Current projects include:
- design of biosensors for minimal polyketides / engineering type-III polyketide product specificity
- engineering E. coli for anaerobic biological activation of short-chain alkanes via fumarate addition
- directed evolution of alkylsuccinate synthase substrate specificity
- engineering ligand specificity of the ItcR repressor for biosensor applications;
In collaboration with other labs, we more broadly develop and apply molecular and synthetic biology tools. For example, we are developing controllable / inducible gene expression systems in human T-cells using an engineered bacterial repressor protein (MarR) (collaborations Navin Varadarajan (UH) and Dmitry Nevozhay (MD Anderson Cancer Center). Working with Jacinta Conrad (UH), we are designing novel, tunable “appendage” mutants of E. coli and M. hydrocarbonoclasticus; these are useful for studying/engineering bacterial adhesion in bioremediation applications, and bacteria-driven emulsions.
We gratefully acknowledge the National Science Foundation for numerous grants to support these research projects.
Graduate student projects and other research details can be found on the Cirino Lab Website.
Current Lab Members:
Graduate Students
- Ehsan Bahrami (current Ph.D. student)
- Nam Nguyen (current Ph.D. student)
- Yixi Wang (current Ph.D. student)
Undergraduate Students
- Tania Pena Reyes (University of Chicago - Rowley Scholarship)
- Ana Alvarez (Volunteer)
- Carson Bush (2021 SURF recipient)
- Joshua Nwose
Former Lab Members:
Postdocs
- Dr. Joseph Gredell (now Senior Scientist at Novozymes)
- Dr. Shuang-Yan Tang (now Professor at Chinese Academy of Sciences, Institute of Microbiology)
- Dr. Hui Li
Graduate Students
- Aarti Doshi (PhD 2020); Research Scientist at Tessera Therapeutics
- Zhiqing Wang (PhD 2019); Research Scientist at Mascoma LLC
- Shuai Qian (PhD 2018); Senior Scientist at Solugen
- Ye Li (PhD 2017); Assistant Professor, Jiangnan University
- Shaza Abnouf (PhD student of N Varadarajan (primary advisor))
- Christopher Frei (PhD 2016); Senior Scientist, Manus Biosynthesis
- Balakrishnan Ramesh (PhD student of N Varadarajan (primary advisor)); Scientist at Muufri, Inc
- Dr. Olubolaji Akinterinwa (PhD 2010): Scientist at Procter & Gamble
- Dr. Jonathan Chin (PhD 2010); Senior Scientist at Algenol Biofuels
- Dr. Reza Khankal (PhD 2010); Glycos Biotechnology then Chevron
- Dr. Hossein Fazelinia (PhD 2009 co-advised by C.Maranas); Children’s Hospital, Philadelphia
- Lexan Lhu (M.S.; Joule Biotechnologies, MA)
- Francesca Luziatelli (visiting PhD student from U of Tuscia, Italy)
- Ryan McLay (co-advised) (graduated M.S. student)
- Lois Eppihimer (lab manager)
Former Undergraduate Researchers
- Tasnuva Haider (volunteer)
- Ibrahim Hassan (volunteer)
- Anthara Krishnan (volunteer)
- Nam Nguyen - SURF
- Priya Patel – SURF
- Zheng You (volunteer)
- Diep Nguyen – SURF
- Jennie Nguyen - SURF
- Aya Elsaadi - SURF
- Rachel Dunn - PURS
- Justin Hood (volunteer)
- Danielle Valcourt (REU, Notre Dame)
- Katy Norman (REU, Trinity)
- Wayne Andrade (UH, volunteer)
- Chelsea Kraynak (University of MD, REU summer 2012)
- Oliver Chou (U Colorado Boulder) REU
- Kyle Schutter (Brown University REU; now CEO of Takamoto Biogas Ltd.)
- Nicholas Linn (NC State) - REU
- Ajay Padaki
- Berook T. Alemayehu (U Maryland: Baltimore County) REU
- Audrey Leung (University of Iowa) REU
- Alex Teng (UC Berkeley) - REU
- Caroline Monroe (Penn State)
- Anthony Tascone (Penn State)
- Renae Patch (Penn State)
- Garrett Tobin (Penn State)
- Jeremy Sargent (Penn State)
- J. Lane Weaver (Penn State)
- Jon P. Badalamenti (Penn State)
- Lucien E Weiss (Penn State)
- Julie Sawlsville (Penn State)
- Geoff Geise (Penn State) - REU
- Doug Haag (Penn State)
- Megan Rex (Penn State)
- Chris Cottle (Penn State)
- Panagiotis A Papadopoulos (Penn State)
- Matthew Zapadka (Penn State)
- Galen Lynch (PSU)
- Noah Johnson (PSU)
- Chris Frei (PSU)
- Trevor Wiley
- Erica Trump (West Virginia U) – REU
- Nimrah Ahmed
- Shariful Alam
- Aubrey Dolly
- Daniel Seong
Work-Study Students
- Joshua Nwose
- Hiba Ahmed
- Chiemela Ubani
- Marilu Serna
- Arlene Sanchez
- Ghazal Nemati
- Christian Miles
- Mulbah Kallen
- Louis Avila
- Zeal Patel
- Areeba Ahmed
- Sana Basheer
- Ryan Noraas
- Tiffany Veet
- Khai Van
- Jon Ajak
2006 N.S.F. CAREER Award Recipient
Associate Editor: BMC Biotechnology
Editorial Board Member: PLOS One
Board of Advisors, Ohio University Dept. of Chemical & Biomolecular Engineering
Guest Editor, AIChE Journal (2020, honoring Frances Arnold)
Guest Editor, Current Opinion in Biotechnology (multiple issues)
Guest Editor, Journal of Biomedicine and Biotechnology (2010)
ACS BIOT 2012 Program Chair
ACS Symposium Chair 2008, 2009; 2014 (CATL); 2015, 2016
SIMB Biocatalysis program co-chair, 2010 - 2014
Proposal Reviewing: NSF, NIH, DOE (BER, BES), ARPA-E, USDA, others...
- Cirino PC and Ingram LO, inventors, Materials and methods for the efficient production of xylitol in E. coli, U.S. Patent Application Serial No. 20070072280. Licensed to BioEnergy InternationalDate: 09/01/2005
- Arnold FH and Cirino PC, inventors. Thermostable peroxide-driven cytochrome P450 oxygenase variants and methods of use. U.S. Patent No. 7,435,570. Licensed to CodexisDate: 08/11/2004
- Cirino PC and Arnold FH, inventors. Peroxide-driven cytochrome P450 oxygenase variants. U.S. Patent No. 7,704,715. Licensed to CodexisDate: 04/16/2002
Selected Publications
- A Doshi, I Bandey, D Nevozhay, N Varadarajan, PC Cirino, Design and characterization of a salicylic acid-inducible gene expression system for Jurkat cells. Journal of Biotechnology 346, 11-14, 2022
- Y Wang, N Nguyen, SH Lee, Q Wang, JA May, R Gonzalez, PC Cirino, Engineering Escherichia coli for anaerobic alkane activation: Biosynthesis of (1‐methylalkyl)succinates. Biotechnology and Bioengineering 119 (1), 315-320, 2022
- A Doshi, F Sadeghi, N Varadarajan, PC Cirino, Small-molecule inducible transcriptional control in mammalian cells. Critical Reviews in Biotechnology 40 (8), 1131-1150, 2020
- Z Wang, A Doshi, R Chowdhury, Y Wang, CD Maranas, PC Cirino, Engineering sensitivity and specificity of AraC-based biosensors responsive to triacetic acid lactone and orsellinic acid. Protein Engineering, Design and Selection 33, 2020
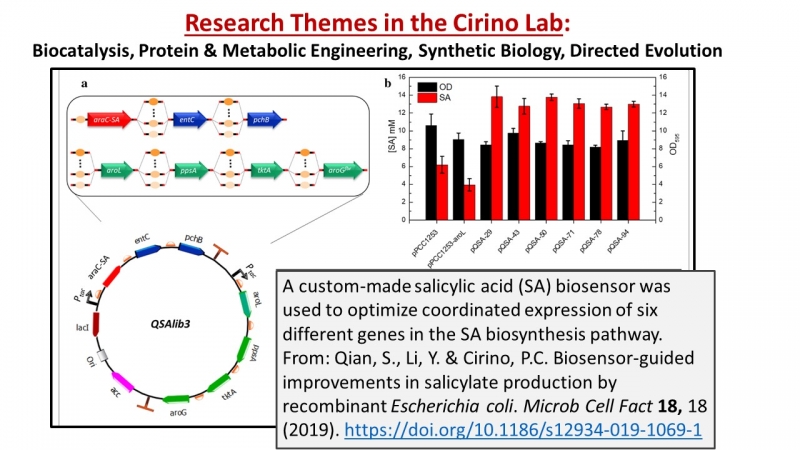
- S Qian, Y Li, PC Cirino, Biosensor-guided improvements in salicylate production by recombinant Escherichia coli. Microbial Cell Factories 18., 2019
- CS Frei, S Qian, PC Cirino, New engineered phenolic biosensors based on the AraC regulatory protein. Protein Engineering, Design and Selection 31 (6), 213-220, 2018
- RB McLay, HN Nguyen, YA Jaimes-Lizcano, NK Dewangan, Simone Alexandrova, Debora F Rodrigues, Patrick C Cirino, Jacinta C Conrad, Level of Fimbriation Alters the Adhesion of Escherichia coli Bacteria to Interfaces. Langmuir 34 (3), 1133-1142, 2018
- Y Li, S Qian, R Dunn, PC Cirino, Engineering Escherichia coli to increase triacetic acid lactone (TAL) production using an optimized TAL sensor-reporter system. Journal of Industrial Microbiology and Biotechnology 45 (9), 789-793, 2018
- Y Li, Z Wang, PC Cirino, Design and characterization of new β-glucuronidase active site variants with altered substrate specificity. Biotechnology letters 40 (1), 111-118, 2018
- Frei CS, Wang ZQ, Qian S, Deutsch S, Sutter M, & Cirino PC, Analysis of amino acid substitutions in AraC variants that respond to triacetic acid lactone. Protein Science, 25(4), 804-814. doi: 10.1002/pro.2873, 2016
- Qian S, & Cirino PC, Using metabolite-responsive gene regulators to improve microbial biosynthesis. Current Opinion in Chemical Engineering, 14, 93-102. doi: 10.1016/j.coche.2016.08.020, 2016
- Sharma S, Jaimes-Lizcano YA, McLay RB, Cirino PC, & Conrad, JC, Subnanometric Roughness Affects the Deposition and Mobile Adhesion of Escherichia coli on Silanized Glass Surfaces. Langmuir, 32(21), 5422-5433. doi: 10.1021/acs.langmuir.6b00883, 2016
- Wang ZQ, & Cirino PC, New and improved tools and methods for enhanced biosynthesis of natural products in microorganisms. Current Opinion in Biotechnology, 42, 159-168. doi: 10.1016/j.copbio.2016.05.003, 2016
- Li Y and PC Cirino, Recent advances in engineering proteins for biocatalysis. Biotechnology & Bioengineering, 111(7), 1273-1287., 2014
- Grisewood MJ, Gifford NP, Pantazes RJ, Li Y, Cirino PC, Janik MJ, and CD Maranas, OptZyme: computational enzyme redesign using transition state analogues. PLoS One.8(10):e75358, 2013
- Qian S and PC Cirino, Protein engineering as an enabling tool for synthetic biology. In Synthetic Biology: Tools and Applications. 23-42. Edited by H. Zhao. Elsevier, 2013
- Tang SY, Qian S, Akinterinwa O, Frei CS, Gredell JA, and PC Cirino, Screening for enhancedtriacetic acid lactone production by recombinant Escherichia coli expressing a designedtriacetic acid lactone reporter. Journal of the American Chemical Society. 135:10099-10103, 2013
- Gredell JA, Frei CS and PC Cirino, Protein and RNA engineering to customize microbial molecular reporting. Biotechnology Journal. 7(4):477-99, 2012
- Ramesh B, Sendra VG, Cirino PC, and N Varadarajan, Single-cell characterization of autotransporter-mediated Escherichia coli surface display of disulfide bond-containing proteins. Journal of Biological Chemistry. 287(46):38580-9., 2012
- Akinterinwa O and PC Cirino, Anaerobic obligatory xylitol production in Escherichia coli strains devoid of native fermentation pathways. Applied and Environmental Microbiology.77(2):706-9, 2011
- Chin JW and PC Cirino, Strain engineering strategies for improving whole-cell biocatalysis: engineering Escherichia coli to overproduce xylitol as an example. Nanoscale Biocatalysis: Methods in Molecular Biology, 743:185-203, 2011
- Chin JW and PC Cirino, Improved NADPH supply for xylitol production by engineered Escherichia coli with glycolytic mutations. Biotechnol. Progress, 27(2):333-41, 2011
- Chin, J. W.; Cirino, P. C., Improved NADPH Supply for Xylitol Production by Engineered Escherichia Coli with Glycolytic Mutations. Biotechnology Progress 2011, 27 (2), 333-341., 2011
- Fasan R, Crook NC, Landwehr M, Cirino PC, and FH Arnold, Improved product-per-glucose yields in P450 dependent propane biotransformations using engineered E. coli. Biotechnol Bioeng., 108(3):500-10, 2011
- Fasan, R.; Crook, N. C.; Peters, M. W.; Meinhold, P.; Buelter, T.; Landwehr, M.; Cirino, P. C.; Arnold, F. H., Improved NADPH supply for xylitol production by engineered Escherichia coli with glycolytic mutations. Biotechnol. Progress, 27(2):333-41, 2011
- Tang SY and Cirino PC, Design and application of a novel mevalonate-responsive regulatory protein. Angewandte Chemie International Edition. 50(5):1084-6, 2011
- Cirino PC, Metabolic engineering strategies for production of commodity and fine chemicals: Escherichia coli as a platform organism. Manual of Industrial Microbiology and Biotechnology. ASM Press, ISBN: 9781555815127, 2010
- Tang SY and PC Cirino, Elucidating residue roles in engineered variants of AraC regulatory protein. Protein Sci. 19(2):291-8, 2010
Editorials / Edited Volumes
- Alper H, Cirino P, Nevoigt E, and Sriram G, Applications of Synthetic Biology in Microbial Biotechnology. J Biomedicine Biotechnology. Article ID 918391, 2020
SELECTED PUBLICATIONS
- Akinterinwa O and PC Cirino, Heterologous expression of D-xylulokinase from Pichia stipitis enables high levels of xylitol production by engineered Escherichia coli growing on xylose. Metab Eng. 11(1):48. Cover article, 2009
- Akinterinwa, O and PC Cirino, Catabolism and Metabolic Fueling Processes. In: The Metabolic Pathway Engineering Handbook, ISBN: 9780849339233, 2009
- Chin JW, Khankal R, Monroe CA, Maranas CD, and PC Cirino, Analysis of NADPH supply during xylitol production by engineered Escherichia coli. Biotechnol. Bioeng. 102(1):209.Spotlight article, 2009
- Fazelinia H, Cirino PC, and CD Maranas, OptGraft: A computational procedure for transferring a binding site onto an existing protein scaffold. Protein Sci. 18(1):180, 2009
- Frei CS and PC Cirino, Combinatorial Enzyme Engineering. In: Protein Engineering and Design. J.R. Cochran and S. Park, Eds. Publisher: CRC; 1 edition, ISBN-10: 1420076582, 2009
- Khankal R, Chin JW, Ghosh D, and PC Cirino, Transcriptional effects of CRP* expression in Escherichia coli. J. Biol. Eng. 3:13, 2009
- Khoury GA, Fazelinia H, Cirino PC, and CD Maranas, Computationally driven redesign of Candida boidinii xylose reductase for altered cofactor specificity. Protein Sci.18(10):2125-38. Cover article, 2009
- Akinterinwa O, Khankal R, and PC Cirino, Metabolic Engineering for Bioproduction of Sugar Alcohols. Curr Opin Biotechnol. 19(5):461, 2008
- Cirino PC and Sun L, Advancing Biocatalysis through Enzyme, Cellular, and Platform Engineering. Biotechnol. Progr. 24(3):515, 2008
- Khankal R, Chin JW, PC Cirino, Role of xylose transporters in Escherichia coli engineered for xylitol production. J. Biotechnol. 134(3-4):246, 2008
- Khankal R, Luziatelli F, Chin JW, Frei CS and PC Cirino, Comparison between three common Escherichia coli lab strains as platforms for xylitol production. Biotechnol Lett.30(9):1645, 2008
- Tang SY, Fazelinia H, and PC Cirino, AraC regulatory protein mutants with altered effector specificity. J. Am. Chem. Soc. 130(15):5267, 2008
- Weiss LE, Badalamenti JP, Weaver JL, Tascone AR, Weiss PS, Richard TL, and PC Cirino, Engineering motility as a phenotypic response to LuxI/R-dependent quorum sensing in Escherichia coli. Biotechnol Bioeng. 100(6):1251. Spotlight article, 2008
- Badalamenti JP, Weiss LE, Buckno CJ, Richard TL, Weiss PS, and PC Cirino, Synthetic Sports: A Bacterial Relay Race. IET Synth. Biol. 1:61, 2007
- Fazelinia H, Cirino PC, and CD Maranas, Extending IPRO in protein library design for ligand specificity. Biophys. J. 92(6):2120, 2007
- Cirino PC, Chin JW and LO Ingram, Engineering Escherichia coli for Xylitol Production from Glucose-Xylose Mixtures. Biotechnol. Bioeng. 95: 1167, 2006
- Cirino PC and FH Arnold, A self-sufficient peroxide-driven hydroxylation biocatalyst. Angew. Chem.-Int. Edit. 42: 3299, 2003
- Cirino PC and R Georgescu, Screening for thermostability. Methods Mol Biol. 230: 117, 2003
- Cirino PC, Mayer KM, and D Umeno, Generating mutant libraries using error-prone PCR. Methods Mol Biol. 231: 3, 2003
- Cirino PC, Tang Y, Takahashi K, Tirrell DA and FH Arnold, Global Incorporation of Norleucine in Place of Methionine in Cytochrome P450 BM-3 Heme Domain Increases Peroxygenase Activity. Biotechnol. Bioeng. 83: 729, 2003
- Salazar O*, Cirino PC* (*equal contributions) and FH Arnold, Thermostabilization of a Cytochrome P450 Peroxygenase. ChemBioChem. 4: 891, 2003
- Cirino PC and FH Arnold, Protein engineering of oxygenases for biocatalysis. Curr. Opin. Chem. Biol. 6: 130, 2002
- Cirino PC and FH Arnold, Regioselectivity and activity of cytochrome P450 BM-3 and mutant F87A in reactions driven by hydrogen peroxide. Adv. Synth. Catal. 344: 932, 2002